- Research
- Open access
- Published:
Association analysis of gut microbiota with LDL-C metabolism and microbial pathogenicity in colorectal cancer patients
Lipids in Health and Disease volume 23, Article number: 367 (2024)
Abstract
Background
Colorectal cancer (CRC) is the most common gastrointestinal malignancy worldwide, with obesity-induced lipid metabolism disorders playing a crucial role in its progression. A complex connection exists between gut microbiota and the development of intestinal tumors through the microbiota metabolite pathway. Metabolic disorders frequently alter the gut microbiome, impairing immune and cellular functions and hastening cancer progression.
Methods
This study thoroughly examined the gut microbiota through 16S rRNA sequencing of fecal samples from 181 CRC patients, integrating preoperative Low-density lipoprotein cholesterol (LDL-C) levels and RNA sequencing data. The study includes a comparison of microbial diversity, differential microbiological analysis, exploration of the associations between microbiota, tumor microenvironment immune cells, and immune genes, enrichment analysis of potential biological functions of microbe-related host genes, and the prediction of LDL-C status through microorganisms.
Results
The analysis revealed that differences in α and β diversity indices of intestinal microbiota in CRC patients were not statistically significant across different LDL-C metabolic states. Patients exhibited varying LDL-C metabolic conditions, leading to a bifurcation of their gut microbiota into two distinct clusters. Patients with LDL-C metabolic irregularities had higher concentrations of twelve gut microbiota, which were linked to various immune cells and immune-related genes, influencing tumor immunity. Under normal LDL-C metabolic conditions, the protective microorganism Anaerostipes_caccae was significantly negatively correlated with the GO Biological Process pathway involved in the negative regulation of the unfolded protein response in the endoplasmic reticulum. Both XGBoost and MLP models, developed using differential gut microbiota, could forecast LDL-C levels in CRC patients biologically.
Conclusions
The intestinal microbiota in CRC patients influences the LDL-C metabolic status. With elevated LDL-C levels, gut microbiota can regulate the function of immune cells and gene expression within the tumor microenvironment, affecting cancer-related pathways and promoting CRC progression. LDL-C and its associated gut microbiota could provide non-invasive markers for clinical evaluation and treatment of CRC patients.
Introduction
According to GLOBOCAN 2020 data from the International Agency for Research on Cancer (IARC), colorectal cancer (CRC) incidence ranks third, following breast and lung cancers, while its mortality rate is second only to lung cancer. Each year, over 1.9 million new CRC cases are predicted globally, with 0.935 million deaths [1]. Social and economic development, coupled with sedentary lifestyles and increased consumption of animal-derived foods, leads to reduced physical activity and obesity, which are independently related to CRC risk [2]. Although the mechanisms underlying CRC are unclear, research shows that obesity and sedentary lifestyles may cause lipid metabolism disorders [3, 4]. Additionally, lipid metabolism disorders are increasingly recognized as important roles in cancer progression, including CRC [5]. Cholesterol, an important component of blood lipids, is highly lipophilic and is transported by lipoproteins, a lipid-protein complex. Low-density lipoprotein cholesterol (LDL-C), a primary type of lipoprotein, transports cholesterol from the liver to various tissues, providing raw materials for tissue cells, including cancer cells [6]. Studies have shown that LDL-C receptor levels are upregulated in CRC patients, and the addition of LDL-C to cell cultures significantly increases ROS levels in CRC cells, alters gene expression, and activates the MAPK pathway, thereby enhancing intestinal tumorigenicity and accelerating tumor progression. [7]. Moreover LDL-C mediates the occurrence of CRC through its oxidation to oxidized low-density lipoprotein (oxLDL) [8]. A similar study revealed that the interaction between LDL-C and the mucin family gene MUC4 rs1104760A > G may be important in diagnosing CRC. This combination may induce CRC by affecting LDL-C levels [9]. Meanwhile, retrospective cross-sectional studies have shown that elevated LDL-C levels are significantly associated with lymph node metastasis in various cancers, including CRC [10].
Primary prevention is crucial in reducing the global burden of CRC. Although endoscopic examinations can reduce CRC incidence and mortality, flexible sigmoidoscopy is ineffective for proximal colon cancer and challenging for large-scale screening due to cost and invasiveness [11]. With deeper understanding of CRC metagenomics, gut microbiota offer new perspectives for CRC diagnosis and therapy. Bacteria such as Parvimonas mira and Solobacterium moorei serve as non-invasive biomarkers for CRC [12, 13], whereas Bacteroides vulgatus and Akkermansia muciniphila exhibit anti-cancer effects on CRC cell proliferation [14]. Additionally, gut microbiota can regulate host metabolism and show promise in studies on lipid metabolism associated with intestinal tumors. For example, P anaerobius enriched in colon tumors and adenoma tissues may interact with toll-like receptors to increase intracellular active oxidants, promoting cholesterol synthesis and CRC cell proliferation [15]. Squalene epoxidase(SQLE), an essential enzyme in cholesterol synthesis, can mediate intestinal tumor occurrence through the gut microbiota-metabolic axis [16].
Thus, LDL-C, a major type of cholesterol, has a close relationship with CRC and interacts with gut microbiota. While research has shown that intestinal microbiota preparations can lower human LDL-C levels [17], no studies have yet suggested a relationship between intestinal microbiota of CRC patients and LDL-C. This study sought to investigate the makeup and abundance of intestinal microbiota in the feces of CRC patients, identify internal relationships among typical gut microbiota and their relationships with elevated LDL-C levels, investigate microbial factors responsible for LDL-C metabolism disorders in CRC patients, and explore possible internal regulation among these gut microorganisms. Subsequent research will focus on the immune and biological mechanisms driven by typical gut microbiota in CRC development amidst irregular LDL-C metabolism, and create predictive models to biologically assess LDL-C levels in CRC patients.
Methods
Participant details and inclusion criteria
The Medical Ethics Committee of the Guangxi Medical University Cancer Hospital has approved this research protocol. All participants signed an informed consent form prior to surgery and were notified about the sampling before sample collection. Based on the inclusion criteria, researchers collected fecal samples collected from 236 CRC patients prior to treatment between January 1, 2021 and December 31, 2021, and ultimately collected 198 fecal samples that passed the 16S ribosomal RNA (16S rRNA) sequencing quality test. Concurrently, freshly collected tissue samples of the aforementioned subjects who underwent surgical treatment at the Guangxi Medical University Cancer Hospital were collected and stored in cryogenic liquid nitrogen. Among them, 181 cancer patients had LDL-C information. Additionally, among the 17 CRC patients who underwent transcriptomic sequencing of tumor tissue samples, LDL-C data were available for 14 samples, and 8 samples simultaneously underwent 16S rRNA sequencing from pre-treatment fecal samples.
The inclusion criteria for this study include: 1. Patients who underwent surgery and have a clear pathological classification (staging in accordance with the ACJJ CRC classification guidelines), or CRC patients diagnosed by colonoscopy histopathological biopsy; 2. No history of comorbidities or other malignant tumors in the past; 3. Excluding other gastrointestinal disorders, there are no acute complications such as complete bowel obstruction and gastrointestinal perforation; 4. Prior to collecting fecal specimens, none of the patients had received any cancer therapy, including surgical procedure, chemo, radiation therapy, immune therapy, and traditional Chinese medicine treatment; 5. Not using antibiotics and gut microbiota preparations within the past month; 6. unconscious disorders or other cognitive impairments.
Collection of stool samples and 16S rRNA sequencing
After receiving notification of the sampling plan, the subjects collected fecal samples on the second day of admission. During the sampling process, members of research group guided the subjects to avoid urine contamination, and used sterile fecal collection tubes to retain the middle part of the fecal sample. Subsequently, the fecal sample was stored in a sterile ice container, encased in a 2 mL EP tube with a dosage of 200 mg per tube, and preserved in a refrigerator at -80° C. Following the dispatch of fecal specimens to the lab with the MOBIO PowerSoil® DNA Isolation Kit, DNA was isolated from 200 mg of feces using Tris–EDTA buffer, adhering to the prescribed product guidelines. Following the extraction of DNA, the specimens undergo DNA quality testing, permitting those of satisfactory quality to advance to the subsequent experiment. Primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) were used to focus on and secure the V3 and V4 segments of the 16S rRNA gene, followed by PCR amplification of these targeted sequences. Post-PCR amplification, the initial analysis of each sample’s PCR products was conducted through 2% agarose gel electrophoresis, aiming for a band size between 300 and 350 base pairs, with a sequencing depth of 50,000 reads to capture the target sequences. Subsequently, the PCR outputs were measured with the Quant-iT PicoGreen dsDNA Assay Kit, and all specimens were merged in equal molar amounts, adhering to the sequencing criteria derived from each sample’s quantitative outcomes. Subsequently, the KAPA Library Quantification Kit KK4824 was employed to measure the quantity of the mixed libraries. Ultimately, sequencing of the libraries was conducted using an Illumina PE250 device by Genesky Biotechnologies company, (Shanghai, China), employing a 2 × 250 bp approach after successful completion of the library preparation.
Tissue sample collection and transcriptome high throughput sequencing
Based on the premise that the interval between separation and storage in liquid nitrogen is within 30 min, fresh tissues, with soybean size, were obtained from surgically removed tumors and adjacent normal tissue. Using Trizol ® The Total RNA Extraction Kit extracted total RNA from 17 CRC tumor samples and detected the integrity of the RNA using electrophoresis. RNA purity was determined through micro ultraviolet spectrophotometers. Refer to the instructions of the RNA seq sample preparation kit (VAHTS ™ Stranded mRNA-seq Library Prep Kit for Illumina ®), remove rRNA and construct cDNA library. The transcriptome library sequencing was performed using the Illumina NovaSeq 6000 by GENE + company, (Beijing, China). The unprocessed sequencing dataset was evaluated for quality by FastQC, and the valid dataset was first compared with the reference genome using HISAT2 (version: hg38). Gene expression was evaluated using StringTie and known gene models, and the calculated TPM (Transcripts Per Million) values were used to quantify the expression abundance of each gene.
Analysis of tumor immune infiltration
Using the ‘CIBERSORT R script v1.03’ through R software, the CIBERSORT algorithm constructs a feature matrix derived from microarray data from tumor tissue sequencing. Subsequently, the TPM matrix was transformed into a relative abundance gene feature matrix of 22 immune cells (including B cells, CD4 + T cells, CD8 + T cells, neutrophils, macrophages, dendritic cells and various varieties and functional statuses of immune cells) for tumor immune infiltration analysis [18].
Functional annotation analysis of transcriptome sequencing related to LDL-C
The Single Sample Gene Set Enrichment Analysis (ssGSEA) [19] algorithm calculates the matrix of gene set scores for each sample using the GSVA software package v1.46.0, based on downloaded gmt format gene set files (c2.cp.kegg.v2022.1.Hs.symbols.gmt, c5.go.v2022.1.Hs.symbols.gmt). Next, the L-LDL-C group was used as the control group, the variations in Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways among the groups were examined using the limma algorithm in the TCGAbiolinks package v2.25.3. GO enrichment analysis encompasses three aspects: biological processes (BP), cellular components (CC) and molecular functions (MF). The threshold for statistical significance of differentially expressed genes is: P < 0.05 and |log2FC|> 0.
Construction and recognition of machine learning models for gut microbiome biomarkers
Using the multilayer perceptron (MLP) model and the XGBoost (XGB) model to identify gut microbiota markers, respectively, to predict LDL-C levels in CRC patients. MLP is a feedforward artificial neural network model, which comprises an input layer, several hidden layers, and an output layer. Employing backpropagation technology, MLP iteratively adjusts the weights between neurons, ultimately achieving the construction of a neural network between the input and output layers [20]. XGB, a boosting-based integration algorithm, uses information from previous trees to enhance the quality of the current tree for iterative generation by building learners in parallel [21]. As a typical integration of classification and regression tree cart algorithms, XGB has improved the traditional Gradient Boosting Decision Tree [22]. These improvements include the introduction of additional regularization, integrated tree pruning, and subsampling features in XGB, which significantly alleviate overfitting problems, as well as the use of techniques for calculating generalized gain scores to simplify optimization problems in boosting trees [23].
Linking the SciKit Learn 0.18(https://scikit-learn.org/stable/) Platform and Python, utilized downloaded installation packages to construct and assess machine learning models. According to a 7:3 ratio, microbiota dataset of 181 patients who met the inclusion criteria was randomly split into training and testing sets. Subsequently, MLP and XBG models were developed and predicted using LDL-C related differential gut microbiota species with differential importance in the top 15%. Finally, the receiver operating curve (ROC) and area under curve (AUC) were applied to assess the models’ accuracy performance.
Analysis method for 16S rRNA sequencing
Qualitative Insights Into Microbiological Ecology version 2 was used to perform quality filtering on the FASTQ raw sequencing data of all samples. Subsequently, species were annotated according to the Greengenes database v13.8, while intestinal microbiota ASV/OTU was extracted using the photoseq package v126.1. Firstly, the gut microbiota diversity within the group was evaluated employing α-diversity, where Chao1 and ACE characterized the species abundance of the microbiota, while Shannon and Simpson described the microbial diversity and evenness. Secondly, β-diversity was used to evaluate the variability of the microbial structure in each sample across distinct groups. ANOSIM and ADONIS analyses were performed employing the vegan package v2.5.6. Subsequently, the mixOmics v6.6.2 software package was employed to complete partial least squares discriminant analysis (PLS-DA). Next, Linear discriminant analysis Effect Size (LEfSe) analysis was performed employing LEfSe software v1.0.0, combined with linear discriminant analysis (LDA) to evaluate analysis results, in order to identify species with significant abundance differences between groups (employing |LDA|> 2 and P < 0.05 as difference screening thresholds). Ultimately, employing PICRUSt2 software 2.3.0 to predict the KEGG pathways enriched between sequencing sample groups, and calling vegan package v2.5.6, the study used non-parametric Mann–Whitney U rank-sum test to analyze the inter group diversity indices and KEGG pathway variability. Finally, the ggplot2 package v3.4.0 was used to visualize histograms. The above operations were all completed using R software v3.5.1.
Statistical methods
Using SPSS software v23.0, continuous data analysis was performed on clinical data using t-tests, while quantitative data analysis was performed using Pearson Chi-square test. The subsequent procedures were completed using R software v4.2.2. Pearson correlation analysis was used to measure the correlation between gut microbiota and immune cell abundance with immune-related genes. The ggcorplot package v0.1.4 was used to perform Spearman correlation analysis to evaluate the correlation between different subgroups of differential gut microbiota, the correlation between differential microbiota and KEGG pathway, and the correlation between intergroup differential gut microbiota and BP and MF projects. Among them, the ggcorplot software package v0.1.4, Igraph software package v1.3.5, and Cytoscope software v3.7.2 were used to visualize the relevant matrices.
Results
Essential information and clinical features of CRC patients classified by LDL-C levels
Following the application of inclusion and exclusion criteria, patients possessing pre-treatment LDL-C information were enrolled and divided into H-LDL-C and L-LDL-C groups according to their preoperative LDL-C levels. The H-LDL-C group included 80 CRC patients with LDL-C values above 3.37 mmol/L(129.62 mg/dL), while the L-LDL-C group included 101 CRC patients whose LDL-C levels at or underneath the maximum threshold of normal values (the reference range for normal values is 0–3.37 mmol/L (129.62 mg/dL)). As Table 1 shows, CRC patients with different LDL-C levels did not differ significantly in age or sex, suggesting balanced and comparable baseline data. Differences in serum triglyceride levels, serum albumin levels, and Body Mass Index were not statistically significant, indicating comparable nutritional status between the two groups. Additionally, patients with H-LDL-C had a higher percentage of abnormal total cholesterol compared to those in the L-LDL-C group (P < 0.001), while high-density lipoprotein cholesterol (HDL-C) did not differ significantly between the two groups, suggesting that LDL-C might be associated with cholesterol metabolism disorders in CRC patients.
Comparison of microbial diversity between H-LDL-C and L-LDL-C groups in CRC Patients
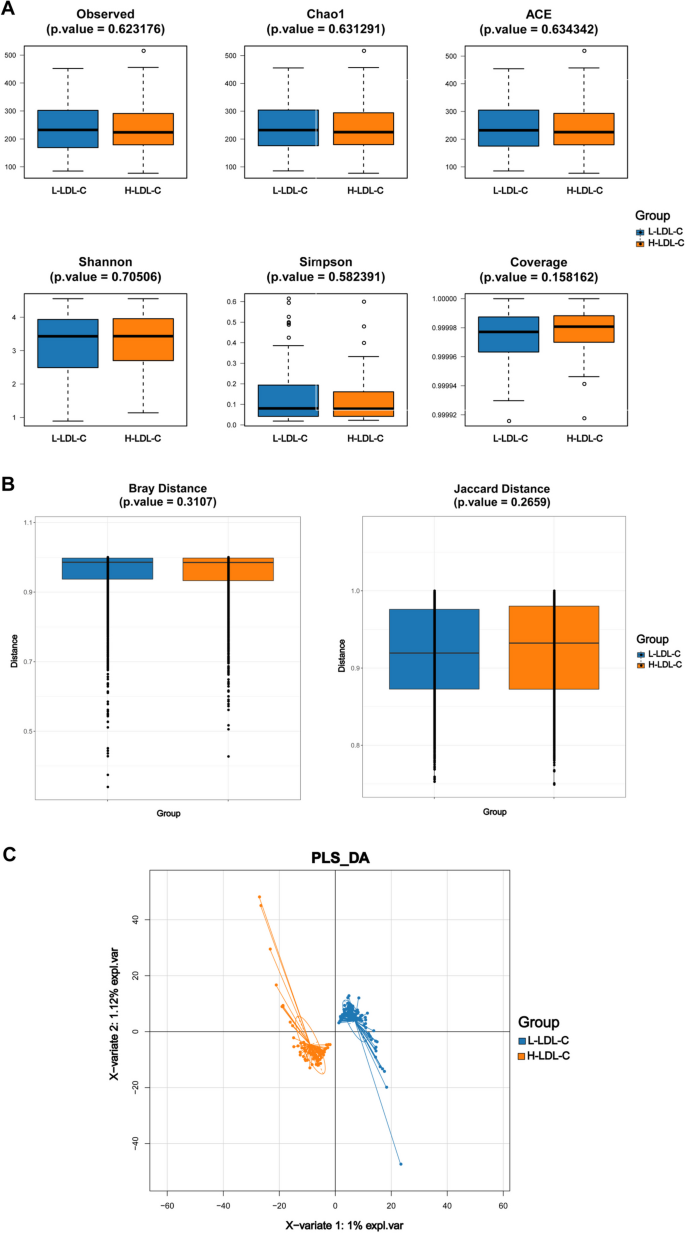
At the start of the study, differences in microbial diversity between the H-LDL-C and L-LDL-C groups of CRC patients were investigated using α-diversity and β-diversity indices. α-diversity for samples from both patient groups is shown in Fig. 1A. Although differences were observed, they were not statistically significant (P > 0.05). Figure 1B presents the β-diversity for two CRC patient groups. Bray (P = 0.3107) and Jaccard (P = 0.2659) indices suggest no statistically significant differences in gut microbiota composition between the groups (Supplementary Tables 1 and 2). PLS-DA analysis, shown in Fig. 1C, revealed that CRC patients in two groups clustered according to their gut microbiota. The findings suggest that although there were no statistically significant differences in fecal microbiota diversity within and between groups, CRC patients persist substantial differences in gut microbiota composition based on LDL-C levels.
Comparison of gut microbiota diversity index between L-LDL-C group and H-LDL-C group patients with CRC. A Comparison of α-diversity index of gut microbiota between L-LDL-C group and H-LDL-C group in CRC patients. B Comparison of β-diversity index of gut microbiota between L-LDL-C group and H-LDL-C group in CRC patients. The horizontal axis represents the group, the vertical axis represents the diversity index value of the sample community within the group, and the color also represents the group. C PLS-DA analysis of gut microbiota in the L-LDL-C and H-LDL-C groups of CRC patients. The dots represent each sample of gut microbiota, the color represents the group, the horizontal and vertical axis scales represent the relative distance of each sample, and X variable 1 and X variable 2 represent the factors that affect the changes in gut microbiota composition of CRC patients in the L-LDL-C and H-LDL-C groups, respectively
Identification of gut microbiota associated with abnormal LDL-C metabolism
To investigate gut microbiota with varying abundance between H-LDL-C and L-LDL-C groups and identify key biomarkers for abnormal LDL-C metabolism, the study performed LEfSe analysis on these two groups. The analysis revealed significant statistical variations in the abundance of 24 microbial communities between two groups. Both H-LDL-C and L-LDL-C groups exhibited significantly greater abundance of 12 microbial communities compared to the other group (P < 0.05; see Supplementary Table 3; Fig. 2A, B). Figure 2B displays LDA scores for top10 in each group of these 24 differential microbial communities after LEfSe analysis (log10 transformed). Higher scores indicate greater significance for these species. Correlations between dominant microbial communities in two groups were plotted (Fig. 2C) to further explore their relationships with LDL-C. Among these, four dominant microbial communities in the L-LDL-C group, f__Veillonellaceae.g__Veillonella, f__Corynebacteriaceae.g__Corynebacterium, f__Bifidobacteriaceae.g__S-cardovia and o__Ac-tinomycetales. f__Corynebacteriaceae, were most closely associated with other nodes. These indicate that these four dominant microbial communities are closely related to other dominant microbial communities. Simultaneously, f__Veillonellaceae.g__Veillonella, a dominant microbial community in the L-LDL-C group showed a negative correlation with f__Coriobacteriaceae.g__Paraeg-gerthella, g__Fusobacterium.s__Fusobacterium_necrophorum, and g__Coprobacillus.s__uncultured_organism in the H-LDL-C group. These results indicate that negative regulatory interactions may occur among the dominant microbial communities.
Analysis of differences in gut microbiota between L-LDL-C group and H-LDL-C group CRC patients. A Evolutionary relationship diagram of LEfSe analysis. The node size represents the species abundance and is directly proportional to the species abundance. Node color represents grouping, and yellow nodes in branches represent species with no significant differences in abundance between groups; the red nodes represent species with significantly higher abundance in the L-LDL-C group, while the green nodes represent species with significantly higher abundance in the H-LDL-C group. Each layer node represents a phylum/class/order/family/genus/species from the inside out, and the annotations for each layer's species markers represent a phylum/class/order/family/genus/species from the outside in. B LDA bar chart based on 16S rRNA gene sequencing. The color of the bar chart represents the group, the horizontal coordinate represents the LDA score (after log10 processing), the vertical coordinate represents the species with significantly higher abundance in the group, and the length of the bar chart represents the size of the LDA score value. C LDL-C related differences in gut microbiota correlation network diagram. Each node represents each species, node color represents group, node size represents the number of edges connected to the node. The larger the node, the more edges connected to the node. The connecting line indicates a significant correlation between the two nodes. The blue line represents Spearman correlation coefficient values below 0 (negative correlation), while Spearman correlation coefficient values above 0 (positive correlation) are represented by the red line. The thicker the red line, the greater the Spearman correlation coefficient between two nodes
Predicting gut microbiota function in H-LDL-C and L-LDL-C groups
Next, PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2) software was used to analyze enriched KEGG pathways among characteristic microbiota in two groups to explore the biological relationship between LDL-C and its related gut microbiota. Among the 180 KEGG pathways analyzed, four pathways exhibited statistically significant differences (P < 0.05). The Hypertrophic Cardiomyopathy pathway (P = 0.047) was abundant in H-LDL-C group, while the Steroid hormone biosynthesis (P = 0.042), Steroid biosynthesis (P = 0.042), and Biosynthesis of siderophore group nonribosomal peptides (P = 0.0499) were significantly more abundant in the L-LDL-C group than in the H-LDL-C group (Supplementary Fig. 1 and Supplementary Table 4). These results suggest that gut microbiota associated with LDL-C are closely linked to lipid metabolism in CRC patients.
Relationship between differential gut microbiota associated with LDL-C and immune cells
Tumor-infiltrating immune cells are those that enter the tumor microenvironment (TME) and interact with it, playing a role in either promoting or inhibiting tumor growth. To investigate the connection between LDL-C-associated intestinal microbiota and tumor-infiltrating immune cells, the study created a bar chart to show the composition of 22 immune cells from 14 CRC patients with LDL-C and RNA sequencing dataset (Fig. 3A). Figure 3A shows that each CRC patient has a unique immune cell composition in the TME. Overall, the H-LDL-C group had high proportions of follicular helper T cells (Tfh) and regulatory T cells (Tregs). Conversely, the L-LDL-C group had a high proportion of plasma cells.
The correlation between LDL-C related gut microbiota and tumor immune infiltrating cells. A Bar chart of relative abundance of immune cells in CRC patients grouped by LDL-C status. Each bar represents a sample, and the vertical coordinates represent the predicted relative abundance values of immune cells. The sum of the relative abundances of all immune cells in a single sample is 1, and each color in the graph corresponds to one type of immune cell. B Heat map of the correlation between dominant microbial communities and immune cell abundance in the H-LDL-C group. C Heat map of the correlation between dominant microbial communities and immune cell abundance in the L-LDL-C group. The horizontal axis represents immune cells, and the vertical axis represents microbiota. In the figure, red represents positive correlation, blue represents negative correlation, color depth represents the magnitude of Pearson correlation coefficient, and color from light to dark represents the value of Pearson correlation coefficient from small to large. The “*” in the figure represents the size of the P-value: none * represents a P-value ≥ 0.05, * represents 0.01 ≤ P < 0.05, * * represents 0.001 ≤ P < 0.01, and * * * represents P < 0.001. D Network diagram showing the correlation between LDL-C related differential gut microbiota and immune cells. Each node represents each gut microbiota or immune cell and the connecting line represents a significant correlation between the two nodes; the blue line indicates that the Pearson correlation coefficient is less than 0 (negative correlation), while the red line indicates that the Pearson correlation coefficient is greater than 0 (positive correlation)
To further examine the relationship between immune cells and LDL-C-associated intestinal microbiota, the study analyzed the connection between 22 immune cells and their dominant microbiota in the two groups. In H-LDL-C group, g__Fusobacterium.s__Fusobacterium_necrophorum was significantly positively correlated with Tregs; g__Oscillibacter.s__uncultured_bacterium, f__Shewanellaceae.g__Shewanella, o__Altero-monadales.f__Shewanellaceae, c__Gamma-proteobacteria.o__Alteromonadales, f__Coriobacteriaceae.g__Paraeggerthella, g__Paraeggerthella.s__Paraeggerthella_hongkongensis were significantly positively correlated with resting NK cells. Among these g__Paraeggerthella.s__Paraeggerthella_hongkongensis was also significantly positively correlated with Tfh and significantly negatively correlated with plasma cells(Fig. 3B, D). In the L-LDL-C group, g__Anaerostipes.s__Anaerostipes_caccae was significantly positively correlated with neutrophils, o__Actinomycetales.f__Corynebacteriaceae and f__Corynebacteriaceae.g__Corynebacterium were significantly negatively correlated with plasma cells; f__Veillonellaceae.g__Veillonella was significantly negatively correlated with monocytes (Fig. 3C, D). In summary, there were significant differences in the proportion of tumor-infiltrating immune cells between CRC patients in two groups. Additionally, several dominant gut microbiota in H-LDL-C group showed significant correlations with immune cells, suggesting that LDL-C-associated gut microbiota may influence CRC progression by regulating immune cell infiltration.
The connection between LDL-C-associated gut microbiota and immune-related genes
The immune system is pivotal in cancer progression. To examine the correlation between LDL-C-related intestinal microbiota and immune function, the study conducted a connection analysis between LDL-C related intestinal microbiota and prevalent immune- associated genes. In H-LDL-C group, the dominant gut microbiota o__Alteromonadales.f__Shewanellaceae, g__Oscillibacter.s__uncultured_bacterium, f__Shewanellaceae.g__Shewanella and c__Gammaproteobacteria.o__Alteromonadales were significantly positively correlated with multiple immune checkpoints (KIR3DL1, LAIR1, CD28, and CD80, etc.) (Fig. 4A), chemokines (CCL7, CXCL3, and CCL3, etc.) (Fig. 4B), immune activation genes (CD80 and CD28) (Supplementary Fig. 2), immunosuppressive genes (HAVCR2) (Supplementary Fig. 3) and chemokine receptors (XCR1) (Supplementary Fig. 4). In L-LDL-C group, the dominant gut microbiota g__Butyricimonas.s__uncultured_bacterium and f__Acidamin-ococcaceae.g__Acidaminococcus demonstrated a significant positive correlation with multiple immune checkpoints (PDCD1LG2, TNFSF14, and HAVCR2, etc.) (Fig. 4C), chemokines (CXCL9, CCL8, CCL7, and CCL5, etc.) (Fig. 4D), immune activating genes (TNSF14, TNFSF13B, KLRK1, and CD28, etc.) (Supplementary Fig. 5), immunosuppressive genes (PDCD1LG2 and HAVCR2, etc.) (Supplementary Fig. 6), and chemokine receptors (XCR1, CCR5, and CCR1) (Supplementary Fig. 7). These results suggest that LDL-C-associated differential gut microbiota may serve a vital function in the regulation of immune-related gene expression and the CRC progression.
Correlation between LDL-C related differential gut microbiota and immune related genes. A Heat map of the correlation between dominant gut microbiota and immune checkpoints in the H-LDL-C group. B Heat map of the correlation between dominant gut microbiota and chemokines in the H-LDL-C group. C Heat map of the correlation between dominant gut microbiota and immune checkpoints in the L-LDL-C group. D Heat map of the correlation between dominant gut microbiota and chemokines in the L-LDL-C group. The horizontal axis represents genes and the vertical axis represents gut microbiota. In the figure, red represents positive correlation, blue represents negative correlation, color depth represents the magnitude of Pearson correlation coefficient, and color from light to dark represents the value of Pearson correlation coefficient from small to large. The “*” in the figure represents the size of the P-value: no * represents P-value ≥ 0.05, * represents 0.01 ≤ P < 0.05, * * represents 0.001 ≤ P < 0.01, and * * * represents P < 0.001
Analysis of differential pathways and their connection with gut microbiota according to LDL-C levels
To further investigate the relationship between regulatory pathways associated with LDL-C and LDL-C related gut microbiota, GO and KEGG were conducted. RNA-seq data obtained from tumor specimens of 8 patients, who also underwent 16S rRNA sequencing of intestinal microbiota were converted into scoring matrices using the ssGSEA method. Figures 5A and B show that the analysis of GO and KEGG pathway score matrices for two groups. 139 GO pathways were significantly upregulated in H-LDL-C group [GOBP_KILLING_OF_CELLS_OF_ANOTHER_ORGANISM (logFC = 0.041, P < 0.001), GOMF_EFFLUX_TRANSMEMBRANE_TRANSPORTER_ACTIVITY (logFC = -0.044, P = 0.009) and GOCC_CILIARY_TIP (logFC = -0.031, P = 0.006), etc.] as well as 2 KEGG pathways were significantly upregulated [KEGG_ALDOSTERONE_REGULATED_SODIUM_REAB-SORPTION (logFC = 0.036, P = 0.014) and KEGG_PANTOTHENATE_AND_COA_BIOSYNTHESIS (logFC = 0.040, P = 0.029) (Supplementary Tables 5 and 6 show KEGG and GO list, respectively). The findings indicate that CRC related to LDL-C metabolism exhibit distinct biological functions.
Identification of LDL-C related differential pathways and correlation between differential pathways and LDL-C related differential gut microbiota. A GO volcano plot of LDL-C related differential expression. B KEGG volcano map of LDL-C related differential expression. The horizontal coordinate represents log2 (fold change), and the further the point is from the center, the greater the differential fold; The vertical coordinate represents -log10 (P-value), and the closer to the top point, the more significant the difference in expression. Each point represents the detected differentially expressed genes, with red indicating upregulated genes, blue indicating downregulated genes, and gray indicating no differentially expressed genes. C Correlation diagram between LDL-C related differential BP, MF pathway and differential gut microbiota. The horizontal coordinate represents microbiota, and the vertical coordinate represents GO labels. In the figure, red represents positive correlation, blue represents negative correlation, color depth represents the magnitude of Spearman correlation coefficient, and color from light to dark represents Spearman correlation coefficient value from small to large. In the figure "×" symbol represents the P-value: "×" represents P value ≥ 0.05, without "×" represents P < 0.05
To further investigate the relationship between LDL-C related genomic functions and differential gut microbiota, the study analyzed the correlation between the colony counts of 24 LDL-C-related microbiota from 8 patients and LDL-C-related BP, MF, and KEGG pathway scoring matrices. Significant correlations were observed between some differential microbiota and specific BP and MF pathways. For instance, in the H-LDL-C group, the upregulated pathway GOMF_CARBOHYDRATE_TRANSMEMBRANE_TRANSPORTER_ACTIVITY exhibited a strong positive correlation with o__Sphingobacteriales.f__Chitinophagaceae (r = 0.9, P < 0.05) and f__Coriobacteriaceae.g__Paraeggerthella (r = 0.86, P < 0.05), while GOMF_SUGAR_TRANSMEM-BRANE_TRANSPORTER_ACTIVITY showed a significant positive correlation with these two microbiota (r = 0.81, P < 0.05; r = 0.71, P < 0.05). In the L-LDL-C group, the upregulated pathway GOBP_NEGATIVE_REGULATION_OF_ENDOPLASMIC_RETICULUM_UNFOLDED_PROTEIN_RESPONSE and g__Anaerostipes.s__Anaerostipes_caccae exhibited a pronounced inverse correlation (r = 0.76, P < 0.05) (Fig. 5C; Supplementary Tables 7 and 8). However, upregulated KEGG pathways in both groups of samples did not show significant correlations with these differential gut microbiota (Supplementary Table 9). These results suggest that LDL-C and its associated differential gut microbiota may influence CRC progression through various potential biological interactions.
Construction of biological predictive models for LDL-C status through differential intestinal microbiota
To further identify intestinal microbiota linked to LDL-C and evaluate their prognostic ability, prediction models using MLP and XGB were constructed based on 24 LDL-C related differential gut microbiota identified through LEfSe analysis.
In the MLP-based LDL-C prediction model, the training cohort confusion matrix (Fig. 6A) indicated that the counts of true negative (TN) and true positive (TP) samples were significantly exceeding those of false negative (FN) and false positive (FP) samples. In the validation cohort (Fig. 6B), the number of TN predictions was similar to FN predictions, while TP predictions were notably higher than FP predictions. The ROC curve analysis revealed a 0.940 AUC value for the training cohort and 0.750 for the validation cohort (Fig. 6C).
The effectiveness evaluation of MLP and XGB prediction models. A The confusion matrix of MLP in the training set. B The confusion matrix of the MLP model in the validation set. D The confusion matrix of the XGB model in the training set. E The confusion matrix of the XGB model in the validation set. The Y-axis represents the predicted results of the model, the X-axis represents the true situation, 1 represents correct prediction, 0 represents incorrect prediction, and the value in the box represents the number of samples. C ROC curves of MLP model training and validation sets. F ROC curves of XGB prediction model training and validation sets. The horizontal axis represents the false positive rate predicted by the model, the vertical axis represents the true positive rate predicted by the model, and the area under the curve represents the AUC value. The higher the AUC value, the higher the diagnostic performance of the model
In the XGB-based LDL-C prediction model, the training cohort of confusion matrix (Fig. 6D) showed a significantly higher number of TN and TP samples compared to FN and FP samples. The counts of TN and TP predictions in the validation cohort were greater than those of FN and FP predictions (Fig. 6E). The XGB model’s ROC curve analysis resulted in 0.978 AUC value for the training cohort and 0.601 for the validation cohort (Fig. 6F).
The findings imply that both models demonstrate varying levels of accuracy in predicting LDL-C status, with XGB showing superior performance in the training cohort, while MLP model demonstrated better validation cohort performance.
Discussion
This study examined differences in gut microbiota between CRC patients in H-LDL-C and L-LDL-C groups. The study employed 16S rRNA sequencing to assess the composition and abundance of intestinal microbiota associated with LDL-C in CRC patients. It identified key microbiota essential for distinguishing LDL-C metabolic disorders and used these findings to examine microbial factors related to LDL-C metabolism disorders, interactions among microbial communities, and the causes of microbial variation in CRC patients. Meanwhile, the study investigated the TME and biological functions, and used immune characteristic analysis to investigate the association between particular intestinal microbiota and CRC. Further analyses were performed to evaluate the biological effects of varying microbiota and LDL-C metabolism on CRC progression.
Although HDL-C and TG levels showed no significant differences between two groups, a higher proportion of patients in the H-LDL-C group had abnormal serum total cholesterol levels. The use of the LDL-C regulatory drug Evolocumab, as demonstrated by Koskinas KC et al. to lower LDL-C concentrations in acute coronary syndrome patients, resulted in significant reductions in total cholesterol levels. This suggests that abnormal LDL-C metabolism may play a crucial role in increasing serum total cholesterol levels in CRC patients [24]. LDL-C can promote CRC cell proliferation by regulating lipid metabolism within CRC cells. Additionally, the connection between high cholesterol levels and increased CRC risk further supports the notion that abnormal LDL-C metabolism may be crucial in CRC development [25, 26]. Dynamic monitoring of LDL-C level changes in suspected and high-risk CRC patients could be valuable for tracking disease progression. When examining abnormal LDL-C metabolism, the comparison of microbial diversity between CRC patients in two groups revealed no significant differences in fecal microbiota diversity within or between groups. Similarly, the research conducted by Fu J and colleagues did not find a correlation between gut microbiota and LDL-C levels [27]. This indicates that research on LDL-C-related microbial diversity may need larger sample sizes. Despite the lack of significant diversity results, in this study, PLS-DA analysis revealed notable intergroup distinguishability in gut microbiota. Although further studies are needed to resolve this contradiction, these results indicate that alterations in gut microbiota are linked to LDL-C metabolism.
Whilst, CRC is linked to alterations in diverse intestinal microbiota, including Fusobacterium nucleatum, Peptostreptococcus stomatis, and other microbiota [13]. Therefore, further analysis was conducted on gut microbiota with significant differences in abundance between CRC patients in two groups.
LEfSe analysis showed that Shewanella had a higher abundance in the H-LDL-C group of CRC patients [28]. Shewanella’s unique fatty acid system can produce various fatty acids with a low melting point, including monounsaturated fatty acids (MUFA) and branched-chain fatty acids (BCFA) [29]. When MUFA was used instead of saturated fatty acids, an increase in MUFA intake led to a synchronous decrease in plasma cholesterol concentration due to lower LDL-C levels [30]. The findings suggest increased levels of Shewanella could mark the onset of gut microbiota self-regulation against abnormal LDL-C levels. Shewanella may act as an antagonistic microbiota and a marker of abnormal LDL-C metabolism in CRC patients, potentially serving as a key indicator for managing LDL-C metabolism in these patients. Lactobacillus delbrueckii, significantly enriched in CRC patients of L-LDL-C group, can reverse elevated levels of various lipids, including LDL-C, caused by Staphylococcus aureus and Escherichia coli. Its ability to regulate host lipids has been confirmed by da Costa WKA et al. [31, 32]. Lactobacillus delbrueckii may regulate LDL-C levels by increasing free fatty acid (FFA) levels, which mediate the redistribution of lipid regulatory pools within liver cells, ultimately leading to lower LDL-C levels [33, 34]. This may explain why LDL-C levels did not increase abnormally in CRC patients enriched with Lactobacillus delbrueckii,suggesting its potential use as a live biotherapeutic agent for managing LDL-C metabolic disorders. Additionally, Veillonella, another differential microbiota, showed higher abundance in the L-LDL-C group and was significantly correlated with seven different microbiota. Veillonella can colonize the intestine under inflammatory conditions and is associated with CRC adenocarcinoma and chemotherapy resistance. It is highly enriched in CRC patients’ proximal colon [35,36,37,38]. Among the Veillonella-associated microbiota, Coprobacillus, the dominant genus in the H-LDL-C group, showed a significant increase in abundance in high-fat diet-fed mice and was positively correlated with serum LDL-C levels, while exhibiting low abundance in CRC patients [39, 40]. Therefore, it is speculated that Veillonella negatively regulates Coprobacillus abundance under CRC conditions, thereby affecting LDL-C metabolism. In a high LDL-C environment, Coprobacillus may affect CRC development through decreasing Veillonella abundance. Although these speculations need further confirmation through wet experiments, the results indicate that interactions among microbiota could play a pivotal role in changes in LDL-C levels and disease progression in CRC patients. Investigating strategies to supplement antagonistic microbiota could provide therapeutic benefits for CRC patients.
Although the findings suggest an effect of microbiota on LDL-C and CRC, the mechanisms by which these LDL-C-associated microbiota influence CRC progression remain unclear. The close relationship between intestinal mucosal immunity and gut microbiota has sparked interest in this study. First, microbiota can accelerate the progression of intestinal diseases through affecting immune environment. For example, affected by a high-fat diet, the gut microbiota can promote KRAS mutation driven intestinal carcinogenesis by influencing Major histocompatibility complex II (MHC II) antigen presentation, thereby mediating immune escape [41]. Conversely, immune environment can also directly affect gut microbiota changes. For instance, a defect in surface receptor TLR5 of flagellin can cause unstable changes in gut microbiota and induce chronic intestinal inflammation [42].
Therefore, immune infiltration analysis was conducted on the two patient groups, revealing that Tfh and Tregs had a proportional advantage in the H-LDL-C group. This characteristic could provide an important foundation for subgroup segmentation in CRC patients undergoing immunotherapy. Research has shown that the oxidized product of LDL-C, oxLDL, can regulate Tregs apoptosis and promote the generation of Tfh by modulating Tregs receptor levels [43, 44]. Integrating the results of this research, it can be inferred that a high LDL-C environment is a critical factor affecting the characteristic proportions of Treg and Tfh in the H-LDL-C group. Targeted regulation of LDL-C, in conjunction with CRC immunotherapy, may improve treatment efficacy for patients. Building on the inferred relationship between LDL-C and TME immune cells, the study examined the association between microbiota and immune cell characteristics. It found that Fusobacterium necrophorum, significantly positively correlated with Tregs in H-LDL-C group, was more abundant in CRC tissue. Moreover, the Fusobacterium genus can suppress T cell proliferation and trigger T cell apoptosis, thereby impairing the ability to eliminate and transform cancer cells, similar to Tregs, which inhibit anti-tumor immune function [45]. Based on the correlation between microbiota abundance and Treg infiltration, along with the consistency between microbiota and Treg function, it can be speculated that in a high LDL-C environment, Fusobacterium necrophorum could recruit Tregs through its metabolites to inhibit the anti-tumor immune response, while creating a favorable environment for its own growth via TME immune changes. Therefore, targeted supplementation of Fusobacterium necrophorum for patients with elevated LDL-C levels may slow tumor progression, aiding in achieving a complete comprehensive treatment process.
Subsequently, immune-related gene association analysis was conducted to further investigate the link between LDL-C-associated gut microbiota and immune gene alterations in CRC patients. Among various immune gene associated microbiota, g_Oscillibacter showed high abundance in healthy individuals compared to CRC patients, while g_Butyricimonas exhibited enrichment in CRC tissues [46, 47]. Both g_Butyricimona and g_Oscillibacter showed a co-directional reduction consistent with LDL-C in a study using lactoferrin to regulate metabolic disorders in obese mice [48]. Meanwhile, g_Oscillibacter can promote white adipose tissue inflammatory response by stimulating macrophages, and intestinal inflammation significantly contributes to cancer transformation and tumor progression [49]. Among multiple immune checkpoints positively correlated with LDL-C associated microbiota, KIR is a key factor in regulating NK cell activity [50]. The immune checkpoint KIR3DL1, a gene in the KIR family, provides significant protection against metastasis and peripheral nerve invasion in CRC adenocarcinoma patients by accumulating with other KIR activation genes in the same family [51]. PD-L1 is a promising candidate for CRC immunotherapy. Both PD-L2 and PD-L1 are critical signals in the T cell proliferation activation co-stimulatory molecule family B7: CD28. The immune checkpoint PDCD1LG2, a PD-L2 coding gene, is primarily expressed in monocytes and macrophages associated with CRC tumors. It may inhibit the development of tertiary lymphoid structures formed by the inflammatory aggregation of immune cells during CRC progression [52,53,54]. In the aforementioned analysis, g_Oscillibacter exhibited a significant positive connection with KIR3DL1, while g_Butyricimonas was significantly positively correlated with PDCD1LG2. Combining previous research findings, in high LDL-C environments, g_Oscillibacter and KIR3DL1 play an antagonistic role in regulating cancer progression, jointly affecting CRC. g_Butyricimonas was positively correlated with the immune processes that inhibit CRC development. These findings further underscore the crucial role of LDL-C-related gut microbiota in CRC development. Additionally, these inferences provide insights into tumor immunotherapy and its efficacy evaluation, guided by g_Oscillibacter and g_Butyricimonas.
Subsequently, the biological role of LDL-C metabolic disorders and related gut microbiota in CRC disease was further explored through gene enrichment analysis. The KEGG pathway for pantothenate and CoA biosynthesis was elevated in H-LDL-C group. Pantothenic acid (also known as vitamin B5), a component of coenzyme A, is key in intracellular lipid metabolism [55]. The pantothenic acid and coenzyme A pathways can mediate T cell metabolic reprogramming via oxidative phosphorylation, enhancing anti-tumor activity [56]. This pathway is strongly related to CRC [57]. Additionally, the GO Biological Process pathway involved in the negative regulation of the unfolded protein response in the endoplasmic reticulum was upregulated in the L-LDL-C group. When the homeostasis of endoplasmic reticulum is disturbed by inflammation, hypoxia, or other stimuli, the unfolded protein response is initiated to restore balance. Failure to restore homeostasis results in cell apoptosis [58]. This adaptive response enhances tumor cell adaptability to hypoxic stress, leading to malignant progression [59]. The key transcription factor XBP1 in this reaction can be activated in tumor associated macrophages, promoting CRC growth and metastasis [60]. LDL-C activates the IRE1 and PERK signaling pathways in endoplasmic reticulum’s unfolded protein response [61]. In this study, the dominant microbiota g__Anaerostipes.s__Anaerostipes_caccae in L-LDL-C group was significantly negatively correlated with the GOBP_NEGATIVE_REGULATION_OF_ENDOPLASMIC_RETICULUM_UNFOLDED_PROTEIN_RESPONSE pathway. Anaerostipes can inhibit tumor growth by enhancing CD8 T cells infiltration into CRC tumor tissue [62]. This indicates that LDL-C and Anaerostipes have a dynamic equilibrium, directly or indirectly interfering with endoplasmic reticulum unfolded protein response, thus influencing CRC progression. However, LDL-C and Anaerostipes only showed a negative correlation after synbiotic supplements in lactating pigs [63]. Further research is needed to confirm this conclusion, suggesting that the combination of LDL-C and Anaerostipes may be a potential CRC screening biomarker.
In recent years, machine learning such as the multilayer perceptron (MLP) model and XGBoost (XGB) model have been applied to predict CRC clinical conditions and biochemical indexes [64, 65]. Although LDL-C status can be obtained through serology examination, research on gut microbiota has deepened, making it increasingly useful for the prevention and diagnosis of CRC, including clinical status recognition [66, 67]. Therefore, MLP and XGB models, combined with differential gut microbiota, were used to predict the LDL-C status of CRC patients. Overall, the confusion matrices of both models indicate some false alarm rates in their predictions. However, the AUC of the ROC curve for MLP-based LDL-C prediction model was greater than 0.7 in both training and validation cohorts. For XGB-based model, it has also demonstrated good predictive performance. The findings suggest that both models possess certain predictive precision, with the MLP-based LDL-C prediction model showing better performance. This model has practical clinical utility to detect LDL-C metabolism in CRC patients through gut microbiota. The construction of these models provides more clinical significance for CRC patients’ gut microbiota, offering biological indicators for clinical evaluation and biological treatment of LDL-C-guided CRC.
This research has some limitations, particularly the lack of a healthy population control, which hinders the comparison of LDL-C metabolic disorders and microbial status between CRC patients and healthy individuals. Future studies will include healthy populations to further investigate the relationship between characteristic microbiota in CRC patients and LDL-C metabolic disorders. However, this research aims to identify microbial factors contributing to abnormal LDL-C metabolism in CRC patients and understand the pathogenesis of these differential microbial communities. Comparing two groups of CRC patients is more effective for identifying microbiota related to LDL-C metabolic disorders within the same cancer background. Interestingly, some characteristic bacterial communities, including Veillonella, can colonize other natural human lumens connected to the gastrointestinal tract [68]. Research on the changes and connections between these privileged sites and intestinal microbiota in CRC progression will provide more accurate and personalized biological basis for diagnosing and treating CRC.
Secondly, although the study has identified the characteristic intestinal microbiota potential in improving LDL-C metabolic disorders and aiding in diagnosing and treating CRC through clinical sample data analysis and previous studies, it is essential to validate these results and inferences through wet experiments in experimental organisms. Further exploration of the mechanisms by which gut microbiota mediate LDL-C metabolic disorders and CRC progression is also necessary. Additionally, the data for this study are sourced from real clinical patients, providing irreplaceable biochemical and gut microbiota results for CRC patients. This approach can significantly reduce errors among subjects and disease characteristics, offering research guidance based on microbiota abundance to explore the connection between microbiota and metabolic disorders of LDL-C in CRC patients.
This study focused on microbial factors related to LDL-C metabolic disorders and microbial pathogenicity within the upper limit of clinical LDL-C normal values. Future research will investigate how gut microbiota differ across various LDL-C levels and their relationship with CRC pathogenicity.
Conclusions
The metabolic status of LDL-C in CRC patients is regulated by gut microbiota. When LDL-C levels are abnormally elevated, gut microbiota can influence immune cell function and immune gene expression within the host TME. This, in turn, affects cancer-related biological pathways and promotes CRC progression. LDL-C and its associated gut microbiota could serve as non-invasive biomarkers for CRC clinical evaluation and treatment.
Data availability
The original contributions presented in the study are included in the article material, further inquiries can be directed to the corresponding authors.
Abbreviations
- 16S rRNA:
-
16S ribosomal RNA
- LDL-C:
-
Low-density lipoprotein cholesterol
- CRC:
-
Colorectal cancer
- oxLDL:
-
Oxidized low-density lipoprotein
- SQLE:
-
Squalene epoxidase
- ssGSEA:
-
Single Sample Gene Set Enrichment Analysis
- MUFA:
-
Monounsaturated fatty acids
- BCFA:
-
Branched chain fatty acids
- FFA:
-
Free fatty acids
- MLP:
-
Multilayer perceptron
- XGB:
-
XGBoost
- PLS-DA:
-
Partial least squares discriminant analysis
- ROC:
-
Receiver operating curve
- AUC:
-
Area under curve
- LDA:
-
Linear discriminant analysis
- LEfSe:
-
LDA Effect Size
- TME:
-
Tumor microenvironment
- Tfh:
-
Follicular helper T cells
- Tregs:
-
Regulatory T cells
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto encyclopedia of Genes and Genomes
- MHC II:
-
Major histocompatibility complex II
- BP:
-
Biological processes
- CC:
-
Cellular components
- MF:
-
Molecular functions
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49.
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–64.
Després JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444(7121):881–7.
Bankoski A, Harris TB, McClain JJ, Brychta RJ, Caserotti P, Chen KY, Berrigan D, Troiano RP, Koster A. Sedentary activity associated with metabolic syndrome independent of physical activity. Diabetes Care. 2011;34(2):497–503.
Chen H, Zheng X, Zong X, Li Z, Li N, Hur J, Fritz CD, Chapman W Jr, Nickel KB, Tipping A, Colditz GA, Giovannucci EL, Olsen MA, et al. Metabolic syndrome, metabolic comorbid conditions and risk of early-onset colorectal cancer. Gut. 2021;70(6):1147–54.
Silvente-Poirot S, Poirot M. Cholesterol epoxide hydrolase and cancer. Curr Opin Pharmacol. 2012;12(6):696–703.
Wang C, Li P, Xuan J, Zhu C, Liu J, Shan L, Du Q, Ren Y, Ye J. Cholesterol Enhances Colorectal Cancer Progression via ROS Elevation and MAPK Signaling Pathway Activation. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;42(2):729–42.
Jiang J, Yan M, Mehta JL, Hu C. Angiogenesis is a link between atherosclerosis and tumorigenesis: role of LOX-1. Cardiovasc Drugs Ther. 2011;25(5):461–8.
Kwon MJ, Lee JY, Kim EJ, Ko EJ, Ryu CS, Cho HJ, Jun HH, Kim JW, Kim NK. Genetic variants of MUC4 are associated with susceptibility to and mortality of colorectal cancer and exhibit synergistic effects with LDL-C levels. PLoS ONE. 2023;18(6): e0287768.
Ghahremanfard F, Mirmohammadkhani M, Shahnazari B, Gholami G, Mehdizadeh J. The Valuable Role of Measuring Serum Lipid Profile in Cancer Progression. Oman Med J. 2015;30(5):353–7.
Schoen RE, Pinsky PF, Weissfeld JL, Yokochi LA, Church T, Laiyemo AO, Bresalier R, Andriole GL, Buys SS, Crawford ED, Fouad MN, Isaacs C, Johnson CC, et al. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N Engl J Med. 2012;366(25):2345–57.
Löwenmark T, Löfgren-Burström A, Zingmark C, Eklöf V, Dahlberg M, Wai SN, Larsson P, Ljuslinder I, Edin S, Palmqvist R. Parvimonas micra as a putative non-invasive faecal biomarker for colorectal cancer. Sci Rep. 2020;10(1):15250.
Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, Wang X, Xu X, Chen N, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–8.
Kang X, Ng SK, Liu C, Lin Y, Zhou Y, Kwong TNY, Ni Y, Lam TYT, Wu WKK, Wei H, Sung JJY, Yu J, Wong SH. Altered gut microbiota of obesity subjects promotes colorectal carcinogenesis in mice. EBioMedicine. 2023;93: 104670.
Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, Chan AWH, Chan FKL, Sung JJY, Yu J. Peptostreptococcus anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology. 2017;152(6):1419-1433.e1415.
Li C, Wang Y, Liu D, Wong CC, Coker OO, Zhang X, Liu C, Zhou Y, Liu Y, Kang W, To KF, Sung JJ, Yu J. Squalene epoxidase drives cancer cell proliferation and promotes gut dysbiosis to accelerate colorectal carcinogenesis. Gut. 2022;71(11):2253–65.
Wu Y, Zhang Q, Ren Y, Ruan Z. Effect of probiotic Lactobacillus on lipid profile: A systematic review and meta-analysis of randomized, controlled trials. PLoS ONE. 2017;12(6): e0178868.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–7.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50.
Albaradei S, Thafar M, Alsaedi A, Van Neste C, Gojobori T, Essack M, Gao X. Machine learning and deep learning methods that use omics data for metastasis prediction. Comput Struct Biotechnol J. 2021;19:5008–18.
Ali H, Ahmed A, Olivos C, Khamis K, Liu J. Mitigating urinary incontinence condition using machine learning. BMC Med Inform Decis Mak. 2022;22(1):243.
Wang Z, Xu C, Liu W, Zhang M, Zou J, Shao M, Feng X, Yang Q, Li W, Shi X, Zang G, Yin C. A clinical prediction model for predicting the risk of liver metastasis from renal cell carcinoma based on machine learning. Front Endocrinol. 2022;13:1083569.
Li K, Yao S, Zhang Z, Cao B, Wilson CM, Kalos D, Kuan PF, Zhu R, Wang X. Efficient gradient boosting for prognostic biomarker discovery. Bioinformatics (Oxford, England). 2022;38(6):1631–8.
Koskinas KC, Windecker S, Pedrazzini G, Mueller C, Cook S, Matter CM, Muller O, Häner J, Gencer B, Crljenica C, Amini P, Deckarm O, Iglesias JF, et al. Evolocumab for Early Reduction of LDL Cholesterol Levels in Patients With Acute Coronary Syndromes (EVOPACS). J Am Coll Cardiol. 2019;74(20):2452–62.
Mayengbam SS, Singh A, Yaduvanshi H, Bhati FK, Deshmukh B, Athavale D, Ramteke PL, Bhat MK. Cholesterol reprograms glucose and lipid metabolism to promote proliferation in colon cancer cells. Cancer & metabolism. 2023;11(1):15.
Yao X, Tian Z. Dyslipidemia and colorectal cancer risk: a meta-analysis of prospective studies. Cancer causes & control : CCC. 2015;26(2):257–68.
Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA, Brandsma E, Marczynska J, Imhann F, Weersma RK, Franke L, Poon TW, Xavier RJ, et al. The Gut Microbiome Contributes to a Substantial Proportion of the Variation in Blood Lipids. Circ Res. 2015;117(9):817–24.
Lemaire ON, Méjean V, Iobbi-Nivol C. The Shewanella genus: ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol Rev. 2020;44(2):155–70.
Wang F, Xiao X, Ou HY, Gai Y, Wang F. Role and regulation of fatty acid biosynthesis in the response of Shewanella piezotolerans WP3 to different temperatures and pressures. J Bacteriol. 2009;191(8):2574–84.
Gill JM, Brown JC, Caslake MJ, Wright DM, Cooney J, Bedford D, Hughes DA, Stanley JC, Packard CJ. Effects of dietary monounsaturated fatty acids on lipoprotein concentrations, compositions, and subfraction distributions and on VLDL apolipoprotein B kinetics: dose-dependent effects on LDL. Am J Clin Nutr. 2003;78(1):47–56.
Evivie SE, Abdelazez A, Li B, Bian X, Li W, Du J, Huo G, Liu F. In vitro Organic Acid Production and In Vivo Food Pathogen Suppression by Probiotic S. thermophilus and L. bulgaricus. Front Microbiol. 2019;10:782.
da Costa WKA, Brandão LR, Martino ME, Garcia EF, Alves AF, de Souza EL, de Souza AJ, Saarela M, Leulier F, Vidal H, Magnani M. Qualification of tropical fruit-derived Lactobacillus plantarum strains as potential probiotics acting on blood glucose and total cholesterol levels in Wistar rats. Food research international (Ottawa, Ont). 2019;124:109–17.
Hou G, Yin J, Wei L, Li R, Peng W, Yuan Y, Huang X, Yin Y. Lactobacillus delbrueckii might lower serum triglyceride levels via colonic microbiota modulation and SCFA-mediated fat metabolism in parenteral tissues of growing-finishing pigs. Frontiers in veterinary science. 2022;9:982349.
Daumerie CM, Woollett LA, Dietschy JM. Fatty acids regulate hepatic low density lipoprotein receptor activity through redistribution of intracellular cholesterol pools. Proc Natl Acad Sci USA. 1992;89(22):10797–801.
Rojas-Tapias DF, Brown EM, Temple ER, Onyekaba MA, Mohamed AMT, Duncan K, Schirmer M, Walker RL, Mayassi T, Pierce KA, Ávila-Pacheco J, Clish CB, Vlamakis H, et al. Inflammation-associated nitrate facilitates ectopic colonization of oral bacterium Veillonella parvula in the intestine. Nat Microbiol. 2022;7(10):1673–85.
Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H, Tameda M, Shiraki K, Ito M, Takei Y, Takase K. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: Terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol Rep. 2016;35(1):325–33.
Deng X, Li Z, Li G, Li B, Jin X, Lyu G. Comparison of Microbiota in Patients Treated by Surgery or Chemotherapy by 16S rRNA Sequencing Reveals Potential Biomarkers for Colorectal Cancer Therapy. Front Microbiol. 2018;9:1607.
Sheng Q, Du H, Cheng X, Cheng X, Tang Y, Pan L, Wang Q, Lin J. Characteristics of fecal gut microbiota in patients with colorectal cancer at different stages and different sites. Oncol Lett. 2019;18(5):4834–44.
Li TT, Huang ZR, Jia RB, Lv XC, Zhao C, Liu B. Spirulina platensis polysaccharides attenuate lipid and carbohydrate metabolism disorder in high-sucrose and high-fat diet-fed rats in association with intestinal microbiota. Food research international (Ottawa, Ont). 2021;147:110530.
Yang J, Li D, Yang Z, Dai W, Feng X, Liu Y, Jiang Y, Li P, Li Y, Tang B, Zhou Q, Qiu C, Zhang C, et al. Establishing high-accuracy biomarkers for colorectal cancer by comparing fecal microbiomes in patients with healthy families. Gut microbes. 2020;11(4):918–29.
Schulz MD, Atay C, Heringer J, Romrig FK, Schwitalla S, Aydin B, Ziegler PK, Varga J, Reindl W, Pommerenke C, Salinas-Riester G, Böck A, Alpert C, et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature. 2014;514(7523):508–12.
Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, González A, Clemente JC, Cullender TC, Barnich N, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12(2):139–52.
Gaddis DE, Padgett LE, Wu R, McSkimming C, Romines V, Taylor AM, McNamara CA, Kronenberg M, Crotty S, Thomas MJ, Sorci-Thomas MG, Hedrick CC. Apolipoprotein AI prevents regulatory to follicular helper T cell switching during atherosclerosis. Nat Commun. 2018;9(1):1095.
Li Q, Wang Y, Li H, Shen G, Hu S. Ox-LDL influences peripheral Th17/Treg balance by modulating Treg apoptosis and Th17 proliferation in atherosclerotic cerebral infarction. Cell Physiol Biochemistry. 2014;33(6):1849–62.
King M, Hurley H, Davidson KR, Dempsey EC, Barron MA, Chan ED, Frey A. The Link between Fusobacteria and Colon Cancer: a Fulminant Example and Review of the Evidence. Immune network. 2020;20(4):e30.
Loke MF, Chua EG, Gan HM, Thulasi K, Wanyiri JW, Thevambiga I, Goh KL, Wong WF, Vadivelu J. Metabolomics and 16S rRNA sequencing of human colorectal cancers and adjacent mucosa. PLoS ONE. 2018;13(12):e0208584.
Wu M, Wu Y, Deng B, Li J, Cao H, Qu Y, Qian X, Zhong G. Isoliquiritigenin decreases the incidence of colitis-associated colorectal cancer by modulating the intestinal microbiota. Oncotarget. 2016;7(51):85318–31.
Li L, Ma C, Hurilebagen, Yuan H, Hu R, Wang W and Weilisi. Effects of lactoferrin on intestinal flora of metabolic disorder mice. BMC Microbiol. 2022;22(1):181.
Gaudino SJ, Singh A, Huang H, Padiadpu J, Jean-Pierre M, Kempen C, Bahadur T, Shiomitsu K, Blumberg R, Shroyer KR, Beyaz S, Shulzhenko N, Morgun A, et al. Intestinal IL-22RA1 signaling regulates intrinsic and systemic lipid and glucose metabolism to alleviate obesity-associated disorders. Nat Commun. 2024;15(1):1597.
Dębska-Zielkowska J, Moszkowska G, Zieliński M, Zielińska H, Dukat-Mazurek A, Trzonkowski P and Stefańska K. KIR receptors as key regulators of NK cells activity in health and disease. Cells. 2021;10(7):1777.
Barani S, Hosseini SV, Ghaderi A. Activating and inhibitory killer cell immunoglobulin like receptors (KIR) genes are involved in an increased susceptibility to colorectal adenocarcinoma and protection against invasion and metastasis. Immunobiology. 2019;224(5):681–6.
Yang Z, Wu G, Zhang X, Gao J, Meng C, Liu Y, Wei Q, Sun L, Wei P, Bai Z, Yao H, Zhang Z. Current progress and future perspectives of neoadjuvant anti-PD-1/PD-L1 therapy for colorectal cancer. Front Immunol. 2022;13:1001444.
Lv J, Jiang Z, Yuan J, Zhuang M, Guan X, Liu H, Yin Y, Ma Y, Liu Z, Wang H, Wang X. Pan-cancer analysis identifies PD-L2 as a tumor promotor in the tumor microenvironment. Front Immunol. 2023;14:1093716.
Masugi Y, Nishihara R, Hamada T, Song M, da Silva A, Kosumi K, Gu M, Shi Y, Li W, Liu L, Nevo D, Inamura K, Cao Y, et al. Tumor PDCD1LG2 (PD-L2) Expression and the Lymphocytic Reaction to Colorectal Cancer. Cancer Immunol Res. 2017;5(11):1046–55.
Naquet P, Kerr EW, Vickers SD, Leonardi R. Regulation of coenzyme A levels by degradation: the ‘Ins and Outs.’ Prog Lipid Res. 2020;78:101028.
St Paul MSS, Han S, Israni-Winger K, Lien SC, Laister RC, Sayad A, Penny S, Amaria RN, Haydu LE, Garcia-Batres CR, Kates M, Mulder DT, Robert-Tissot C, Gold MJ, Tran CW, Elford AR, Nguyen LT, Pugh TJ, Pinto DM, Wargo JA, Ohashi PS. Coenzyme A fuels T cell anti-tumor immunity. Cell Metab. 2021;33(12):2415–27.
Yi Y, Wang J, Liang C, Ren C, Lian X, Han C, Sun W. LC-MS-based serum metabolomics analysis for the screening and monitoring of colorectal cancer. Front Oncol. 2023;13:1173424.
Bravo R, Parra V, Gatica D, Rodriguez AE, Torrealba N, Paredes F, Wang ZV, Zorzano A, Hill JA, Jaimovich E, Quest AF, Lavandero S. Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration. Int Rev Cell Mol Biol. 2013;301:215–90.
Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24(19):3470–81.
Zhao Y, Zhang W, Huo M, Wang P, Liu X, Wang Y, Li Y, Zhou Z, Xu N, Zhu H. XBP1 regulates the protumoral function of tumor-associated macrophages in human colorectal cancer. Signal Transduct Target Ther. 2021;6(1):357.
Guevara-Olaya L, Chimal-Vega B, Castañeda-Sánchez CY, López-Cossio LY, Pulido-Capiz A, Galindo-Hernández O, Díaz-Molina R, Ruiz Esparza-Cisneros J and García-González V. LDL promotes disorders in β-cell cholesterol metabolism, implications on insulin cellular communication mediated by EVs. Metabolites. 2022;12(8):754.
Montalban-Arques A, Katkeviciute E, Busenhart P, Bircher A, Wirbel J, Zeller G, Morsy Y, Borsig L, Glaus Garzon JF, Müller A, Arnold IC, Artola-Boran M, Krauthammer M, et al. Commensal Clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe. 2021;29(10):1573-1588.e1577.
Ma C, Gao Q, Zhang W, Zhu Q, Tang W, Blachier F, Ding H, Kong X. Supplementing Synbiotic in Sows; Diets Modifies Beneficially Blood Parameters and Colonic Microbiota Composition and Metabolic Activity in Suckling Piglets. Frontiers in veterinary science. 2020;7:575685.
Qin L, Liang Z, Xie J, Ye G, Guan P, Huang Y, Li X. Development and validation of machine learning models for postoperative venous thromboembolism prediction in colorectal cancer inpatients: a retrospective study. Journal of gastrointestinal oncology. 2023;14(1):220–32.
Du G, Ren C, Wang J, Ma J. The Clinical Value of Blood miR-654-5p, miR-126, miR-10b, and miR-144 in the Diagnosis of Colorectal Cancer. Comput Math Methods Med. 2022;2022:8225966.
Coker OO, Liu C, Wu WKK, Wong SH, Jia W, Sung JJY, Yu J. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome. 2022;10(1):35.
Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene. 2020;39(26):4925–43.
Giacomini JJ, Torres-Morales J, Dewhirst FE, Borisy GG, Mark Welch JL. Site Specialization of Human Oral Veillonella Species. Microbiology spectrum. 2023;11(1):e0404222.
Acknowledgements
None.
Funding
Natural Science Foundation of Guangxi Province (Guangxi Natural Science Foundation) (2021GXNSFAA196008); Youth Science Foundation of Guangxi Medical University (GXMUYSF202402); Youth Research Project of Guangxi Medical University Affiliated Cancer Hospital (yuanqingji2023-10hao); Middle-aged and Young Teachers’ Basic Ability Promotion Project of Guangxi (Basic Ability Promotion Project of Guangxi) (2021KY0087); China Postdoctoral Science Foundation (2023MD734155); Youth Science Foundation of Guangxi Medical University (GXMUYSF202357). Guangxi Key Laboratory of Basic and Translational Research for Colorectal Cancer. Chinese National Natural Science Foundation (82460610).
Author information
Authors and Affiliations
Contributions
M. Q, Y. H, Z.H, X.H, J.L, W. T, X.M: conceived and designed the experiments; J.L, X.H, M. Q, Y.H, Z. H, C.C, Z.W, F.H, C.L, Y. W, B.T, X.M, W.T: analyzed the data; J.L, X.H, M.Q, Z.W, C. C, Y.H, Z.H, F. H, Y. W, C. L, B.T, X.M, W.T: helped with reagents/materials/analysis tools; J.L, M.Q, Y.H, Z. H, X.H, C. C, Z.W, F.H, Y.W, C. L, B.T, X.M, W.T: contributed to the writing of the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics and Human Subject Committee of Guangxi Medical University Cancer Hospital.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
12944_2024_2333_MOESM1_ESM.pdf
Supplementary Material 1: Supplementary Fig. 1. Box plot of KEGG functional abundance in the H-LDL-C group versus the L-LDL-C group of CRC patients. Horizontal coordinates indicate groupings, vertical coordinates indicate predicted abundance values for that pathway in each sample, boxes indicate the 25th-75th percentiles, and the center marker indicates the median; black bars are 1.5 times the interquartile range.
12944_2024_2333_MOESM2_ESM.pdf
Supplementary Material 2: Supplementary Fig. 2. Heat map of the correlation between the dominant flora of H-LDL-C group and immune activation genes.
12944_2024_2333_MOESM3_ESM.pdf
Supplementary Material 3: Supplementary Fig. 3. Heat map of the correlation between the dominant flora of H-LDL-C group and Immunosuppressive genes.
12944_2024_2333_MOESM4_ESM.pdf
Supplementary Material 4: Supplementary Fig. 4. Heat map of the correlation between the dominant flora of H-LDL-C group and chemokine receptors.
12944_2024_2333_MOESM5_ESM.pdf
Supplementary Material 5: Supplementary Fig. 5. Heat map of the correlation between the dominant flora of L-LDL-C group and immune activation genes.
12944_2024_2333_MOESM6_ESM.pdf
Supplementary Material 6: Supplementary Fig. 6. Heat map of the correlation between the dominant flora of L-LDL-C group and Immunosuppressive genes.
12944_2024_2333_MOESM7_ESM.pdf
Supplementary Material 7: Supplementary Fig. 7. Heat map of the correlation between the dominant flora of L-LDL-C group and chemokine receptors. Horizontal coordinates are genes, vertical coordinates are colonies, red in the graph represents positive correlation, blue represents negative correlation, color depth represents Pearson correlation coefficient size, color from light to dark indicates Pearson correlation coefficient value from small to large. The “*” in the graph indicates the size of P value: no * for P value ≥ 0.05, * for 0.01 ≤ P < 0.05, ** for 0.001 ≤ P < 0.01, *** for P < 0.001.
12944_2024_2333_MOESM8_ESM.docx
Supplementary Material 8: Supplementary Table 1. ADONIS test for Bray Distance of intestinal flora in CRC patients in the H-LDL-C and L-LDL-C groups.
12944_2024_2333_MOESM9_ESM.docx
Supplementary Material 9: Supplementary Table 2. ADONIS test for Jaccard Distance of intestinal flora in CRC patients in the H-LDL-C and L-LDL-C groups. Group row: between-group statistics; Residuals row: within-group statistics; Total row: between-group + within-group statistics; Df: degrees of freedom, between-group degrees of freedom are the number of groups—1, within-group degrees of freedom are the total number of samples—number of groups; Sums Of Sqs: sum of squared deviations; Mean Sqs: mean square, the ratio of sums of squared deviations to degrees of freedom, i.e. Sums Of Sqs/ Df; F.Model: F-test value, i.e. between-group mean square/within-group mean square; R2: ratio of between- and within-group sums of squared deviations to total sums of squared deviations, indicating the degree of explanation of differences between samples, with larger R2 indicating a higher degree of explanation of differences between samples; Pr(> F): statistically significant P-value obtained from the substitution test, with Pr < 0.05 as statistically different.
12944_2024_2333_MOESM10_ESM.docx
Supplementary Material 10: Supplementary Table 3. Results of LEfSe analysis. Taxonomy: information of differential species; Group: group with significant abundance of differential species; LDA: effect value of differential species; the table shows species with LDA score (log10) greater than the preset value (default is 2) and P value less than 0.05.
12944_2024_2333_MOESM11_ESM.docx
Supplementary Material 11: Supplementary Table 4. KEGG functional pathways in the intestinal microbiome of CRC patients in the H-LDL-C and L-LDL-C groups. KEGG_Pathway: KEGG pathway; Mean In H-LDL-C: the predicted abundance value of this pathway in each sample in the H-LDL-C group; Mean In L-LDL-C: the predicted abundance value of this pathway in each sample in the L-LDL-C group.
12944_2024_2333_MOESM12_ESM.docx
Supplementary Material 12: Supplementary Table 5. List of differential KEGG pathways of CRC patients stratified by LDL-C condition. FC in logFC is fold change, which indicates the ratio of H-LDL-C group to L-LDL-C expression (and takes the logarithm of its base at 2), with P-value < 0.05 as statistically significant difference.
12944_2024_2333_MOESM13_ESM.docx
Supplementary Material 13: Supplementary Table 6. List of differential GO items of CRC patients stratified by LDL-C condition. FC in logFC, i.e. fold change, denotes the ratio of H-LDL-C group to L-LDL-C expression and was taken as logarithm with a base of 2. P-value < 0.05 was taken as statistically significant difference.
12944_2024_2333_MOESM14_ESM.docx
Supplementary Material 14: Supplementary Table 7. Correclation between enrich GO items of CRC patients in H-LDL-C group and LDL-C-associated differential intestinal microbiome.
12944_2024_2333_MOESM15_ESM.docx
Supplementary Material 15: Supplementary Table 8. Correlation between enriched GO items of CRC patients in L-LDL-C group and LDL-C-associated differential intestinal microbiome.
12944_2024_2333_MOESM16_ESM.docx
Supplementary Material 16: Supplementary Table 9. Correlation between KEGG pathways of CRC patients and LDL-C-associated differential intestinal microbiome. The r.value is the Spearman correlation coefficient value, with P-value < 0.05 as statistically significant difference.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Qin, M., Huang, Z., Huang, Y. et al. Association analysis of gut microbiota with LDL-C metabolism and microbial pathogenicity in colorectal cancer patients. Lipids Health Dis 23, 367 (2024). https://doiorg.publicaciones.saludcastillayleon.es/10.1186/s12944-024-02333-4
Received:
Accepted:
Published:
DOI: https://doiorg.publicaciones.saludcastillayleon.es/10.1186/s12944-024-02333-4